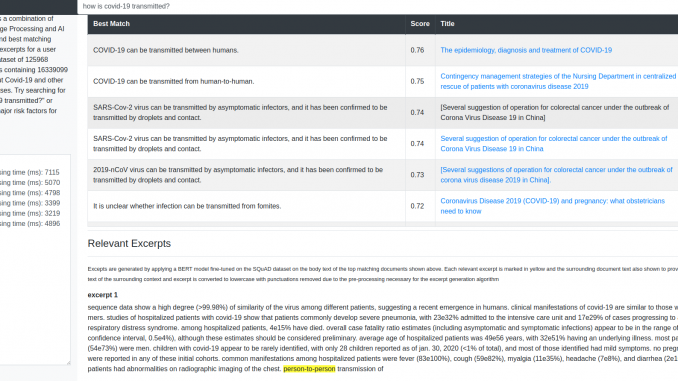
In these series of posts, I’ll describe how I created an Information Retrieval system based on the documents released as part of the Covid-19 research challenge. The system can be accessed here. Try queries such as “how long does covid-19 stay active on plastic surfaces?”, “which country has the highest number of covid-19 cases?”. There are two categories of results. First category is the top-n matching sentences and the documents to which those sentences belong. These are shown in the table at the top of the page. Second category is relevant excerpts from the body text of the top-n documents. These excerpts and surrounding document text are displayed below the table. The excerpts are marked in yellow.
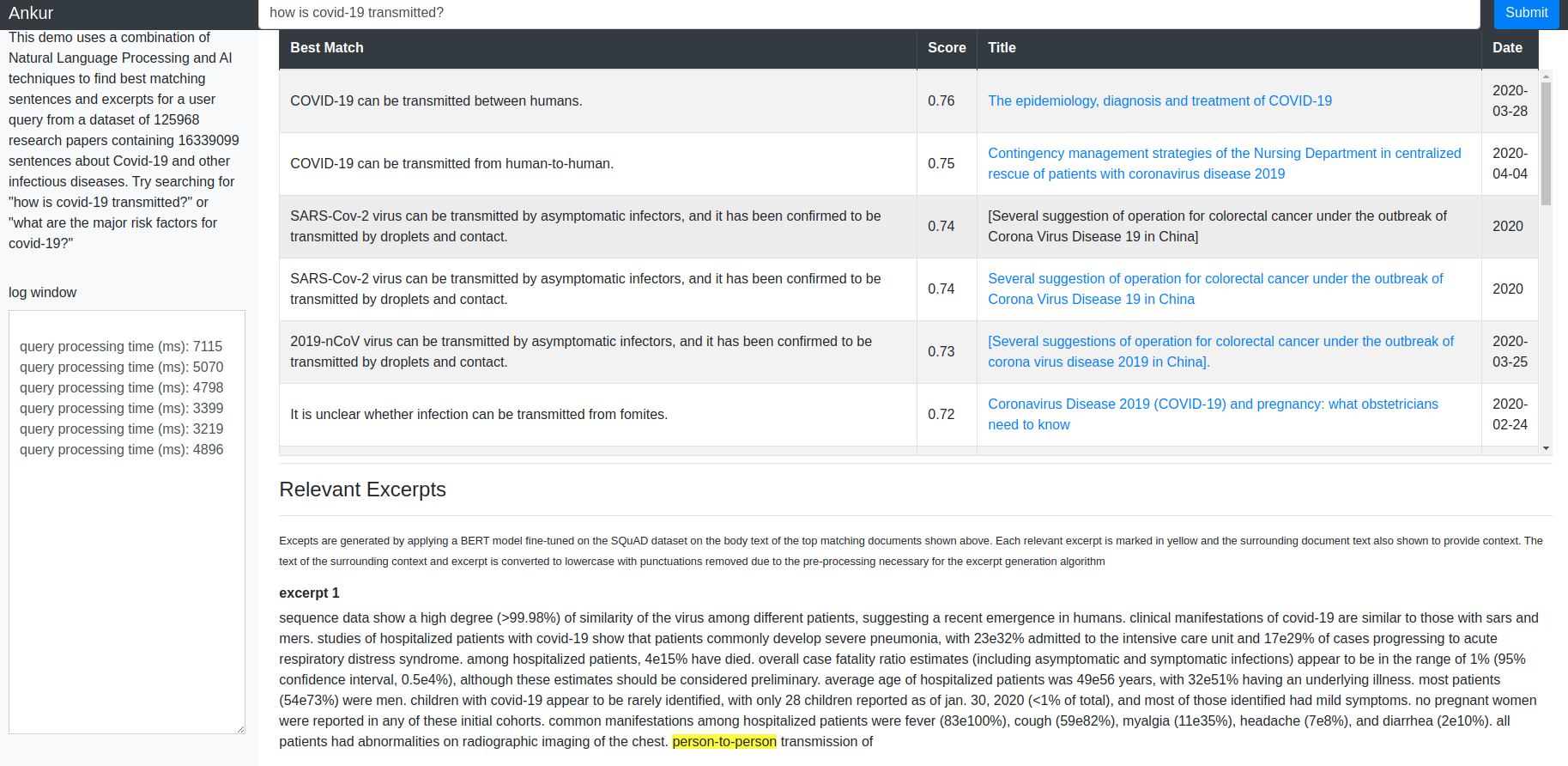
In this post, I’ll go through the ETL (Extract Transform Load) steps needed to parse the document corpus and store document metadata and body text in tables in a SQLite database so that sentence level information can be efficiently queried. I used the following repo for the ETL code and thank the repo author David Mezzitti for making this code available. I learnt a lot going through the code and understanding how it works. Hereafter, I’ll refer to the ETL process as “cord19q ETL” process.
In the next post, I’ll describe how this parsed information is used to create word embeddings, BM25 statistics and sentence embeddings. These sentence embeddings can be used to do a top-n lookup for closest matching sentences and corresponding articles for a user query.
In the third and final post of this series, I’ll go over how the body text of the top-n matching documents can be used as input to a BERT model fine-tuned on a Question-Answering dataset to identify relevant excerpts.
Table of Contents
1. Data Organization
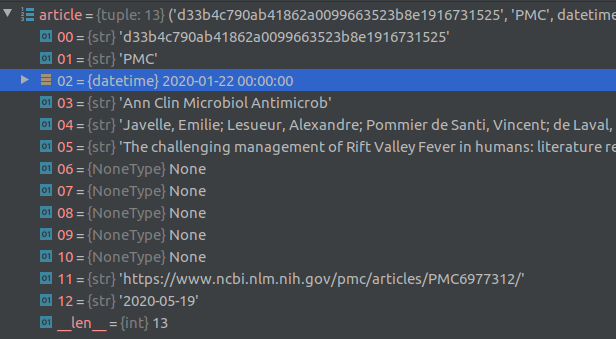
The top level index for the data is a csv file named metadata.csv. Each row in this csv file contains metadata about a paper in the dataset such as the paper title, authors, abstract, date published etc., along with relative links to where the full text of the paper is located. The index to each paper is supposed to be the cord_uid field, however there are a few duplicate cord_uid’s in the csv file (eg., 1g2mup0k). Each row contains a sha value that also serves as the filename of the json file containing the body text of each article. There are a few duplicate sha’s as well, for example: d1dde1df11f93e8eae0d0b467cd0455afdc5b98c, which corresponds to the paper titled “Knowledge synthesis from 100 million biomedical documents augments the deep expression profiling of coronavirus receptors”. This paper appears twice in the list.
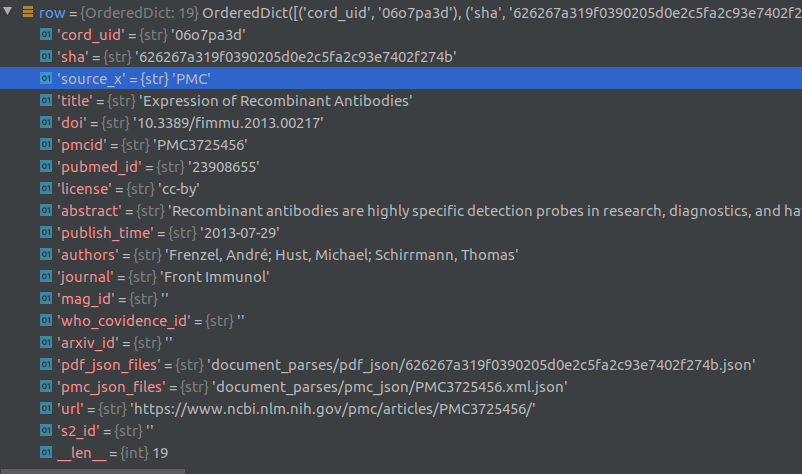
Article full text is contained in json files located in the document_parses/pdf_json and document_parses/pmc_json folders. The relative location of each article’s json file is provided in the pmc_json_files and pdf_json_files columns in the metadata.csv file (see debugger screenshot above). For some articles, there are two corresponding json files, one in the pdf_json folder and the other in the pmc_json folder. It is not clear to me why this is the case.
For about the half the articles, the full text is not available. For such articles, the sha field is empty. See screenshot of part of the metadata.csv file for an example of empty sha fields.

Out of 128162 total number of articles, full text is only available for 55749 articles.
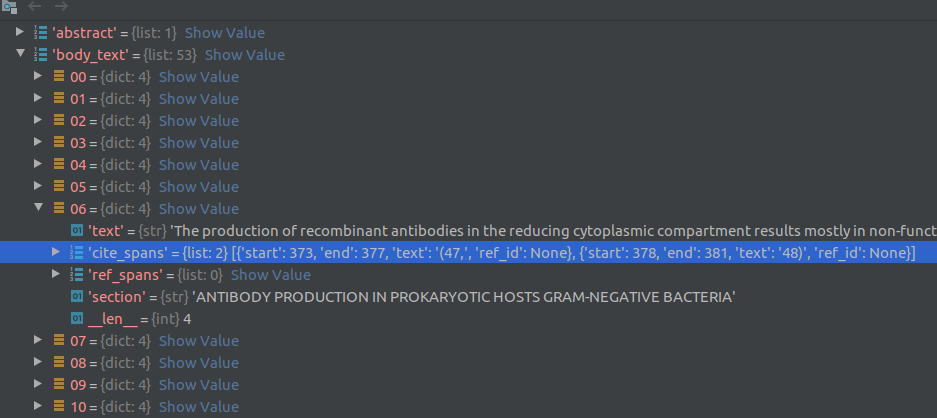
Each json file contains metadata about the article such as article title, authors, abstracts, publication, data published and article text. The article bodytext is a contained in a list of dictionaries (see screenshot below) where each element of the list corresponds to a paragraph of text. A dictionary element contains a string of sentences in the paragraph, the document section of this paragraph (eg., Introduction, Results, Previous work) and cite_spans and ref_spans lists. These lists indicate the starting and ending position of citation and reference marks in the text. This information can be used to clean reference/citation marks (eg., [4], [5]) from the text. Note that the cord19q ETL system uses regular expressions to clean out the citation and reference marks rather than the cite_spans and ref_spans.
Database schemas
The goal of ETL process is to parse the article metadata and body text and store the parsed info in SQLite tables. Four tables are created for this purpose, namely Sections, Articles, Citations and Stats. Out of these 4, the Articles and Sections tables are the most important and contain article and sentence level information respectively. The schemas for these tables is shown below:
|
1 2 3 4 5 6 7 8 9 10 |
# Section table contains sentence level info SECTIONS = { 'Id': 'INTEGER PRIMARY KEY', 'Article': id of the article this sentence comes from 'Tags': Whether this sentence is a fragment or a question (described later) 'Design': Ignore for now 'Name': Section of the sentence (Introduction, References, Results etc) 'Text': Sentence 'Labels': Ignore for now } |
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
# Articles table contains article level info ARTICLES = { 'Id': 'TEXT PRIMARY KEY', 'Source': Article source 'Published':Date published 'Publication': Name of publication where this article appears 'Authors': Article authors 'Title': 'title 'Tags': Article tag - whether this article is relevant to Covid-19 or not. Details described later 'Design': Ignore for now 'Size': Ignore for now 'Sample': Ignore for now 'Method': Ignore for now 'Reference': Article URL } |
Notice that the Article field in the Sections table acts as the foreign key. It cross-references the Sections and Articles tables because it references the Article Id (also called the doc id henceforth) which is the primary key of the Articles table, thereby establishing a link between them.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
# Ignore for now STATS = { 'Id': 'INTEGER PRIMARY KEY', 'Article': 'TEXT', 'Section': 'INTEGER', 'Name': 'TEXT', 'Value': 'TEXT' } # Contains the number of times a given paper title is referenced in citations of other articles CITATIONS = { 'Title': 'TEXT PRIMARY KEY', 'Mentions': 'INTEGER' } |
2. Document parsing workflow
The flowchart below shows the process of creating the Articles and Sections table starting from reading the metadata.csv file.
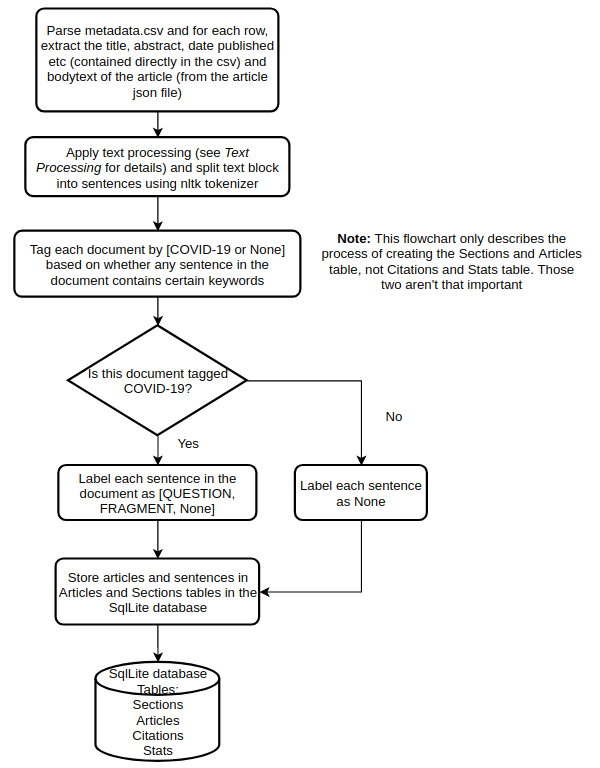
I’ll now go over these steps in more detail.
2.1. Text Processing
For each row in the metadata csv file, the text processing module reads the paper title, abstract, date published and other information present directly in the row columns. For the paper body text, it loads the corresponding json file that contains the full text for that article. The article body text is a described in a list of dictionaries (see screenshot below) where each element of the list corresponds to a paragraph of text. A dictionary element contains a string of sentences in the paragraph, the document section of this paragraph (eg., Introduction, Results, Previous work) and cite_spans and ref_spans lists. These lists indicate the starting and ending position of citation and reference marks in the text.

For each paragraph, the following text cleaning steps are applied:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 |
# Remove emails patterns.append(r"\w+@\w+(\.[a-z]{2,})+") # Remove urls patterns.append(r"http(s)?\:\/\/\S+") # Remove single characters repeated at least 3 times (ex. j o u r n a l) patterns.append(r"(^|\s)(\w\s+){3,}") # Remove citations references (ex. [3] [4] [5]) patterns.append(r"(\[\d+\]\,?\s?){3,}(\.|\,)?") # Remove citations references (ex. [3, 4, 5]) patterns.append(r"\[[\d\,\s]+\]") # Remove citations references (ex. (NUM1) repeated at least 3 times with whitespace patterns.append(r"(\(\d+\)\s){3,}") |
Then, the text processing module uses the nltk sentence_tokenizer to split the text into sentences and creates (section, sentence) tuples as shown in the figure below. The text processing module also attempts to extract text out of any tables in the document. Two screenshots of the Pycharm debugger showing a few (section, sentence) tuples are shown below.
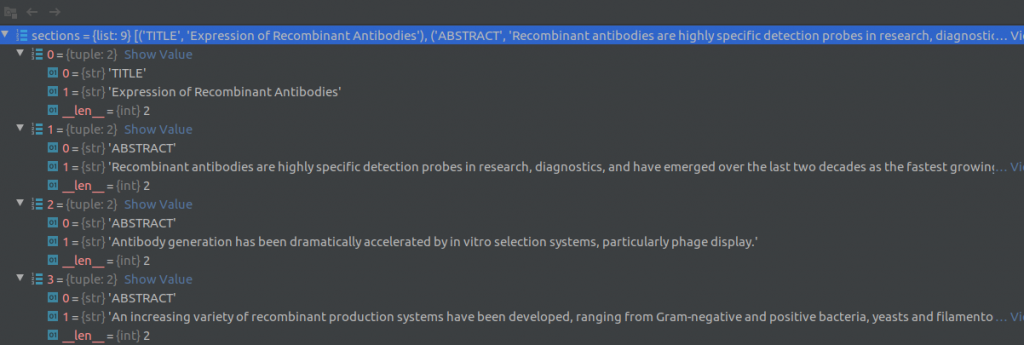
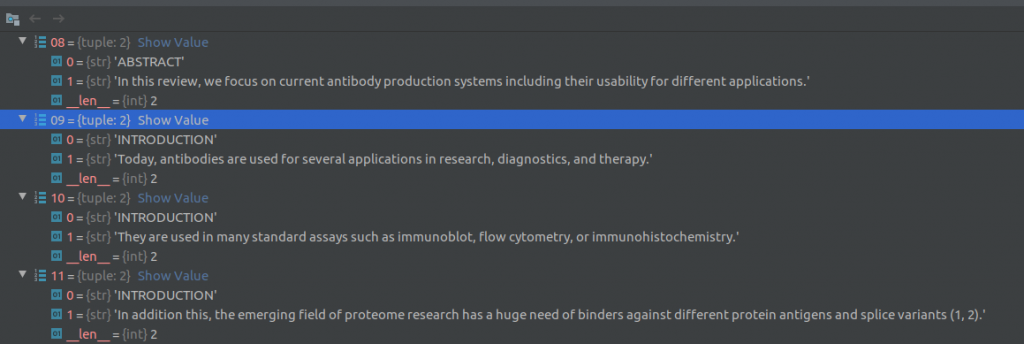
Next, duplicate sentences and sentences that contain “boiler-plate text” are removed. The following phrases constitute boiler-plate text:
|
1 2 |
# Boilerplate text to ignore boilerplate = ["COVID-19 resource centre", "permission to make all its COVID", "WHO COVID database"] |
I’m unclear why this boiler-plate text removal step is necessary.
2.2. Tagging with the COVID-19 tag
If any sentence in a document contains one or more of the following keywords:
|
1 2 |
keywords = [r"2019[\-\s]?n[\-\s]?cov", "2019 novel coronavirus", "coronavirus 2019", r"coronavirus disease (?:20)?19", r"covid(?:[\-\s]?19)?", r"n\s?cov[\-\s]?2019", r"sars-cov-?2", r"wuhan (?:coronavirus|cov|pneumonia)"] |
then that document is tagged with the COVID-19 tag. This tag is used in the downstream preprocessing steps to identify relevant documents. Note that this is a document, not a sentence level tag.
2.3. Tagging Sentences
Next, for the docs tagged with the COVID-19 tag, the ETL system uses the tokenizer and POS (Parts of Speech) tagger in spaCy’s “en_core_sci_md” pipeline to split sentences into list of words and tag each word by its POS tag.
![]()
These tags are used to classify each sentence into Question, Fragment or None (Valid) categories, according to the following rules.
- Questions are sentences that end with a question mark
- Valid sentences satisfy the following rules:
1234# Valid sentences take the following form:# - At least one nominal subject noun/proper noun AND# - At least one action/verb AND# - At least 5 words - All other sentences are fragments.
For example, the sentence “They are almost identical to each other and share 79.5% sequence identify to SARS-CoV.” is a Fragment, because it doesn’t contain a verb.
Note that this classification is only performed for sentences that belong to documents tagged with the COVID-19 tag.
3. Storing parsed information in SQLite tables
The steps described above are executed in parallel for all articles in the dataset by using the Python multiprocessing module. The consolidated output is stored in the Sections and Articles table. The article sha is used as the primary key for the Articles table and a sentence count is used as the primary key for the Sections table. As mentioned before, the Sections table also stores the article sha, which serves as the foreign key to link the Sections and Articles tables.
This concludes this post. In the next post of this series, I’ll describe how this parsed information is used to create word embeddings, BM25 statistics and sentence embeddings. These sentence embeddings can be used to do a top-n lookup for closest matching sentences and corresponding articles for a user query.
Appendix
One pattern you’ll see over and over again in the cord19q code is the use of Python’s multiprocessing module to parallelize the execution of a function across multiple input values, distributing the input data across processes. For example, the ETL process distributes processing the rows of the metadata.csv file across multiple processes. Each process reads the json file corresponding to the article referenced in the row, parses the text as described above and returns back the parsed data to the main process, where the data is inserted into the SQLite database tables.
Note that use of processes instead of threads side steps the Global Interpreter Lock allowing the program to fully leverage multiple processors on a given machine.
I was not familiar with this pattern and found it quite powerful, but also full of pitfalls if one is not careful. Below, I’ll provide two examples of this pattern and highlight a few interesting points.
The code below distributes out the task of squaring a list of numbers (from 0 to 15) to 4 processes by using the pool.imap function. imap applies the function provided as the first argument to the items of the iterable provided in the second argument (it also takes an optional third argument called chunk size which is left to its default value of 1). imap will iterate over the iterable one element at a time, and distribute them across the worker processes, so each worker process works on a separate data item. This is why the result of the code below will be a list of squares of numbers from 0 to 15, and not four copies of each squared value. There are many variants of imap such as map and imap_unordered with small differences. This stackoverflow question provides a better overview than the official multiprocessing documentation.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 |
import os from multiprocessing import Pool def gen(n): for i in range(0, n): yield i def f(x): return x * x if __name__ == "__main__": pool = Pool(processes=4) for i in pool.imap(f, range(16)): print(i) for i in pool.imap(f, gen(16)): print(i) |
In the second example, we’ll do something a bit more useful. Our stream function now will read each row of a metadata.csv file and the multiprocessing module will distribute it out to the process function executed on separate worker processes. In process we simply tokenize the abstract text and return the number of tokens, document sha and process id to the calling process. The calling process accumulates the number of tokens and maintains a map of process ids and the document sha they process.
You can run the program as follows:
|
1 |
python streaming.py 10 |
Here 10 is the number of processes. The program assumes that metadata.csv is in the same directory as the streaming.py file.
The output of this command looks like this on my computer:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 |
processing time: 10.22263479232788 number of unique ids: 55748 word count: 9381582 5668 0 1 2 0 0 0 0 0 0 0 5593 0 0 0 0 0 0 0 0 1 0 5691 1 0 0 0 0 0 0 2 0 1 5518 0 0 0 0 0 0 0 0 0 0 5733 0 0 0 0 0 0 0 0 0 0 5381 0 0 0 0 0 0 0 0 0 0 5737 0 0 0 0 0 0 0 0 0 0 5476 0 0 0 0 0 0 0 0 0 0 5562 0 0 0 0 0 0 0 0 0 0 5392 |
If I run it with 1 as the number of processes, the processing time is 22.5 seconds, so using multiple processes speeds up processing by ~50%.
The matrix shows the overlap between the sha’s processed by the 10 processes. Python’s multiprocessing module takes care of distributing out the rows uniformly across the 10 processes, so each process executes nearly one tenth of the total number of sha’s and there is very little to no overlap between the sets of sha’s processed by each process. In fact, the reason the overlap isn’t always zero is because there are a few duplicate sha’s (as mentioned briefly above).
Lastly, note that each process returns the number of tokens in its lot of documents to the main process where this number is accumulated across all processes. Creating a global count variable and having each process update the count wouldn’t work, because each process gets a copy of the global variables, and the state of these variables isn’t automatically shared. To share state across processes, you must use the Value or Array classes in the multiprocessing module. Generally, sharing state across processes should be avoided. Python multiprocessing works best when the process function takes a small amount of data as input, applies some computationally intensive operations to the data that take a non-trivial amount of time, and returns back a small amount of output. This approach achieves good scaling performance because the overhead of passing data back and forth across processes is kept small and compute cores are utilized efficiently.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 |
import os import csv from multiprocessing import Pool from nltk import sent_tokenize import time import sys curr_dir = os.path.dirname(os.path.abspath(__file__)) def stream(file): ''' :param file: filename (csv file containing article info in this example) :return: row of a csv file ''' with open(os.path.join(curr_dir, file), mode="r") as csvfile: for row in csv.DictReader(csvfile): yield (row) def process(row): ''' :param row: row of a csv file :return: corresponding sha, process id and word count of article abstract ''' abstract = row.get('abstract') sha = row.get('sha') if not sha: return None, None, None sentences = sent_tokenize(abstract) wc = 0 for sentence in sentences: for token in sentence.split(" "): wc = wc + 1 return sha, os.getpid(), wc if __name__ == "__main__": nproc = min(int(sys.argv[1]), os.cpu_count()) ids = set() pid_sha = {} start = time.time() wc = 0 with Pool(nproc) as pool: # use multiple processes to work on rows of the csv file. Python's multiprocessing module ensures # that each process gets its own set of rows to work on, eliminating duplicated work for sha, pid, wc_ in pool.imap(process, stream('metadata.csv')): if sha is None: continue # maintain a dictionary of (process id, [sha]), where the value is a list of sha's that the process id # worked on. That way, we can find out if there are any overlapping sha's. if pid_sha.get(pid) is None: pid_sha[pid] = [sha] else: l_ = pid_sha.get(pid) l_.append(sha) pid_sha[pid] = l_ if sha not in ids: # Skip rows with ids that have already been processed ids.add(sha) wc = wc + wc_ print('processing time: {0}'.format(time.time() - start)) print('number of unique ids: {0}'.format(len(ids))) print('word count: {0}'.format(wc)) # print number of overlaps for k1, v1 in pid_sha.items(): str_ = " " for k2, v2 in pid_sha.items(): overlap = len(list(set(v1) & set(v2))) str_ = str_ + str(overlap) + " " print(str_) print('done') |

Very insightful blogpost. Thank you